
 |

[Mission] [Background] [Core Tools] [Update Center] [Products] [Example Projects] [SourceForge Project Page] [Download] [In Memoriam]
Bio-SPICE, an open source framework and software toolset for Systems Biology, is intended to assist biological researchers in the modeling and simulation of spatio-temporal processes in living cells. In addition, our goal is to develop and serve a user community committed to using, extending, and exploiting these tools to further our knowledge of biological processes.
In collaboration with other Bio-SPICE Community members, we will develop, license, distribute, and maintain a comprehensive software environment that integrates a suite of analytical, simulation, and visualization tools and services to aid biological researchers engaged in building computable descriptions of cellular functions. From disparate data analysis and information mining to experimental validation of computational models of cell systems, our environment will offer a comprehensive substrate for efficient research, collaboration, and publication.
| MODULE | INSTITUTION |
| 2DGrapher | SRI |
| Add Column to Timeseries | UTK |
| Biodata Viewer | SRI |
| BioGrid | UTK |
| BioMat Bridge | SRI |
| BioNets | UNC |
| BioPack | VT |
| BioSens | UCSB |
| BioSketchPad | U Penn/BBN |
| BioSmokey | UTK |
| BioSpreadsheet | UTK |
| BioWarehouse | SRI |
| BioWarehouse Query | SRI |
| BioWarehouse2SBML | Harvard |
| BioWave | NYU |
| Cellx | Indiana |
| Charon | U Penn |
| Clone Updater | TJU |
| CoBi | CFDRC |
| Convert Data to Graph | UTK |
| DBAgent | SRI |
| ESS | UTK |
| Fluxor Computational Analyzer | Harvard |
| Fluxor Spreadsheet | Harvard |
| FTF | |
| GCMConverter | |
| GCMMerger | |
| GeneCite | WRAIR |
| GeneScreen | UCLA |
| Geneways | Columbia |
| Get Column from Timeseries | UTK |
| Get Rows from Timeseries | UTK |
| Graphviz | LBL |
| Graph Viewer | UTK |
| Homologue Finder | LBL |
| Hybrid Automata Symbolic Reachability Tool | Stanford |
| IcDNA | UCLA |
| Jdesigner | KGI |
| Jigcell | VT |
| JournalMine ActionQuery | Columbia |
| Karyote Cell Analyzer (KCA) | Indiana |
| Karyote Genome Analyzer (KAGAN) | Indiana |
| Merge Timeseries Columns | UTK |
| Merge Timeseries Rows | UTK |
| MetaCluster | TJU |
| MetaTool | KGI |
| MIAMESpice | UCLA |
| Model Builder Analyzer | |
| Monod | MSI |
| NCA Analyzer | UCLA |
| NYUMAD | NYU |
| NYUSIM | NYU |
| Octave Bridge | UTK |
| Oscill8 Tools | |
| PAINT | TJU |
| PathwayBuilder | LBL |
| PathwayScreen | WRAIR |
| QIS | QIS |
| PlotML Translator | SRI |
| PtPlot | SRI |
| Reactome | |
| Render Matlab Simulation | LBL |
| Render SBML | LBL |
| SAL | SRI |
| Sample Timeseries | UTK |
| SBML TO Graphviz Dot | LBL |
| SBML to PathwayBuilder | LBL |
| SBML to laid-out Graphviz DOT | LBL |
| SBML:SCC to PathwayBuilder | LBL |
| SBWMatlab | KGI |
| SCCFinder | LBL |
| Sensitivity Analyzer | LBL |
| Simpathica | NYU |
| SOS Tools | Cal Tech |
| Symbolic Analysis Toolbox | Stanford |
| Tab Dlimited Text Converter | SRI |
| TableView | SRI |
| Timeseries to Text | UTK |
| TimeSeries To ZipFile Converter | SRI |
| Vibrio | U Penn |
Experimental systems being studied using Bio-SPICE software include:
|
|
Using comparative genomics software included as part of the Bio-SPICE software, researchers have characterized core elements and variable elements (environment specific) across more than 280 spore forming species; include Bacillus subtilis and Bacillus anthracis. This has provided a starting point for dynamic network analysis, which was performed to classify and analyze feedback loops, identify subnetworks (motifs) that could be potential control points, and analyze dynamic parameterized models. Analysis ranges from elementary modules to full system level, with robust identification of system level control points. Bio-SPICE software modules are used to identify relevant proteins and interactions and use literature searches to filter pathways of interest. Importantly, the computational model has been validated experimentally.

Bio-SPICE analysis of key switches in sporulation network
Overall sporulation network: B. subtilis - common across mutants
Source: Bio-SPICE projects at LBNL (PI: Adam Arkin)
Bio-SPICE has been used to generate an integrated model of metabolic and regulatory network for Myobacterium tuberculosis (Mtb). The stringent response is modeled by a stochastic hybrid system with integrated metabolic and regulatory pathways. The model was verified experimentally both in vivo and in vitro. Boundaries of the flux model kinetic parameters have been described. The Rel gene has been identified as a critical point, with the products regulated by this gene found to be diagnostic markers and therapeutic targets. The developed model has been expanded to simulate dormancy in other pathogens.

Source: Bio-SPICE projects at Univ. of Penn. (PI: Harvey Rubin)
This project is developing tools for metabolic analysis and employing integrated Bio-SPICE to study how mutant organisms optimize their growth rate or minimize their metabolic adjustment relative to the wild-type organism. What are the gaps in our knowledge of the metabolism of this organism? And how can we re-optimize it for new (biotech) goals?
The modeling questions include: Given a genome how do we reconstruct metabolism? How do we generate optimal models? How do we compute the metabolic capabilities of an in-silico representation of an organism? How do we choose a metric for minimizing a mutant organism's metabolic adjustment?
How do we integrate predictions with experimental design and data?
Metabolic models from this pipeline can be used for any annotated microbial genome, e.g., pathogenic Escherichia, Pseudomonas, Yersinia, Bacillus and others of concern to the DoD. In addition, the models could be used to engineer the production and delivery of biomolecules.

Bio-SPICE Minimization of metabolic adjustment - MOMA
Source: Bio-SPICE projects at Harvard (PI: George Church)
In the fight against the HIV virus, the elucidation of retroviruses has been key in fighting the disease. The lack of current computational models also makes HIV a difficult and logical target for malicious engineering. Cell models have been developed to design therapeutic retroviruses and predict parameters critical to the design of these retroviruses. In HIV, the TAT gene has been heavily implicated in whether HIV is latent or active. A higher TAT level results in an increased level of mRNA for the viral genome. The influence of TAT is though to be even more relevant, with measurements obscured by noise.
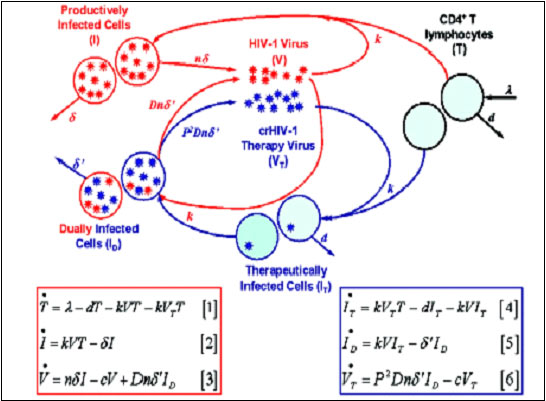
Source: Bio-SPICE projects at LBNL (PI : Adam Arkin)
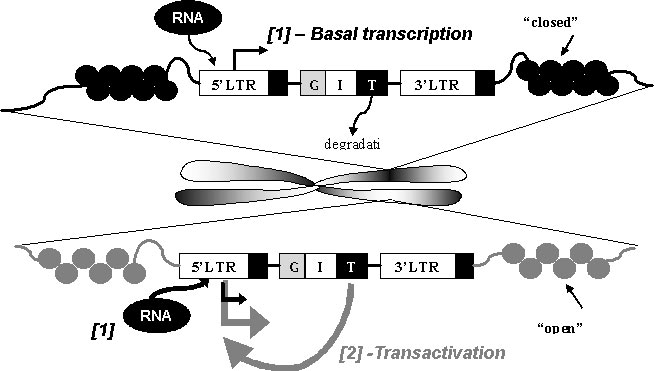
Source: Bio-SPICE projects at LBNL (PI : Adam Arkin)
A key Bio-SPICE discovery found that cell cycle control could be stopped after passing what was conventionally assumed to be the point of no return. A critical checkpoint was identified that allows cell cycle to be arrested and restarted through control of the cyclin pathway. The discovery was made in yeast cells, but a homologous pathway was identified in frog cells, cells which are more complex with a higher level of differentiation. The homologous pathways consisted of similar interactions, but utilized different genes and proteins. The computational model developed contained three modules: an oscillatory module, based on a delayed negative feedback loop; a switching module, based on cooperative removal; and a proteolysis module, based on irreversible activation. Cell cycle arrest was predicted and experimentally verified. Discoveries such as these open new vistas of research, including bacterial growth control, wound-healing, and nerve regeneration.
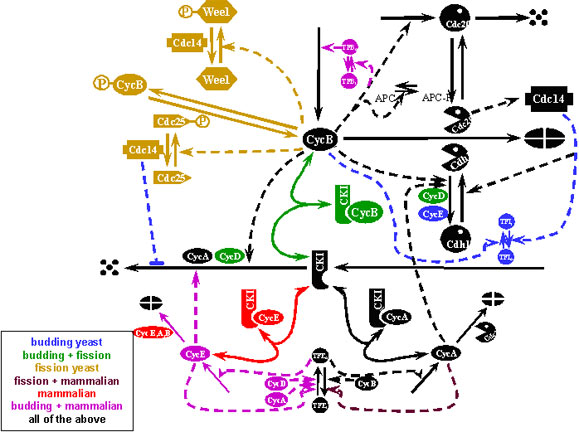
Bio-SPICE assisting rapid validation of CDK control across organisms
Cell cycle control: generic model of CDK control developed
Source: Bio-SPICE projects at VaTech (PI: John Tyson)
This project is developing models and tools for predicting the spatial pattern on hair formation in drosophila wings and cell differentiation in xenopus. Novel stochastic hybrid system models (combining continuous and discrete dynamics) of protein concentrations in a network of cells are developed to characterize hair pattern formation and the dependence on cellular network connectivity and initial conditions. Models use robust parameter identification techniques, symbolic computation and reachability analysis, and ordinary and partial differential equations to represent the system. The role of delta-notch protein concentration dynamics in xenopus differentiation, and the role of Fz, Dsh, and Pk proteins in drosophila wing hair pattern formation are being studied.

Bio-SPICE Spatial Models of networks of cells
Source: Bio-SPICE projects at Stanford Univ. (PI: Claire Tomlin) and at UC Berkeley (PI: Shankar Sastry)
Neutrophil chemotaxis has been elucidated through three dimensional modeling and simulation, allowing reconstruction of the cell shape during location through measurements of polarity and chemical localization. Bio-SPICE offers the unique ability to perform three dimensional simulation of multi-physical systems with one, two, and three dimensional diffusion and changing boundary morphology. The simulation model takes into account both surface and volume measurements, using partial differential equations with stochastic events to model the spatial chemical activity.
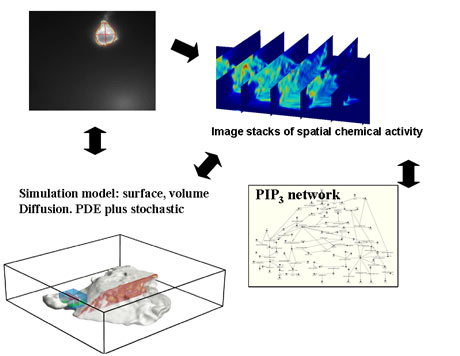
Spatial modeling and simulation essential in many systems
Source: Bio-SPICE projects at LBNL (PI: Adam Arkin)
| The Bio-SPICE project is dedicated to the memory of our friend and colleague, Charles "John" Pedersen. John enthusiastically ran the Bio-SPICE program at SRI during its five years as a DARPA project. We miss you John! |  |